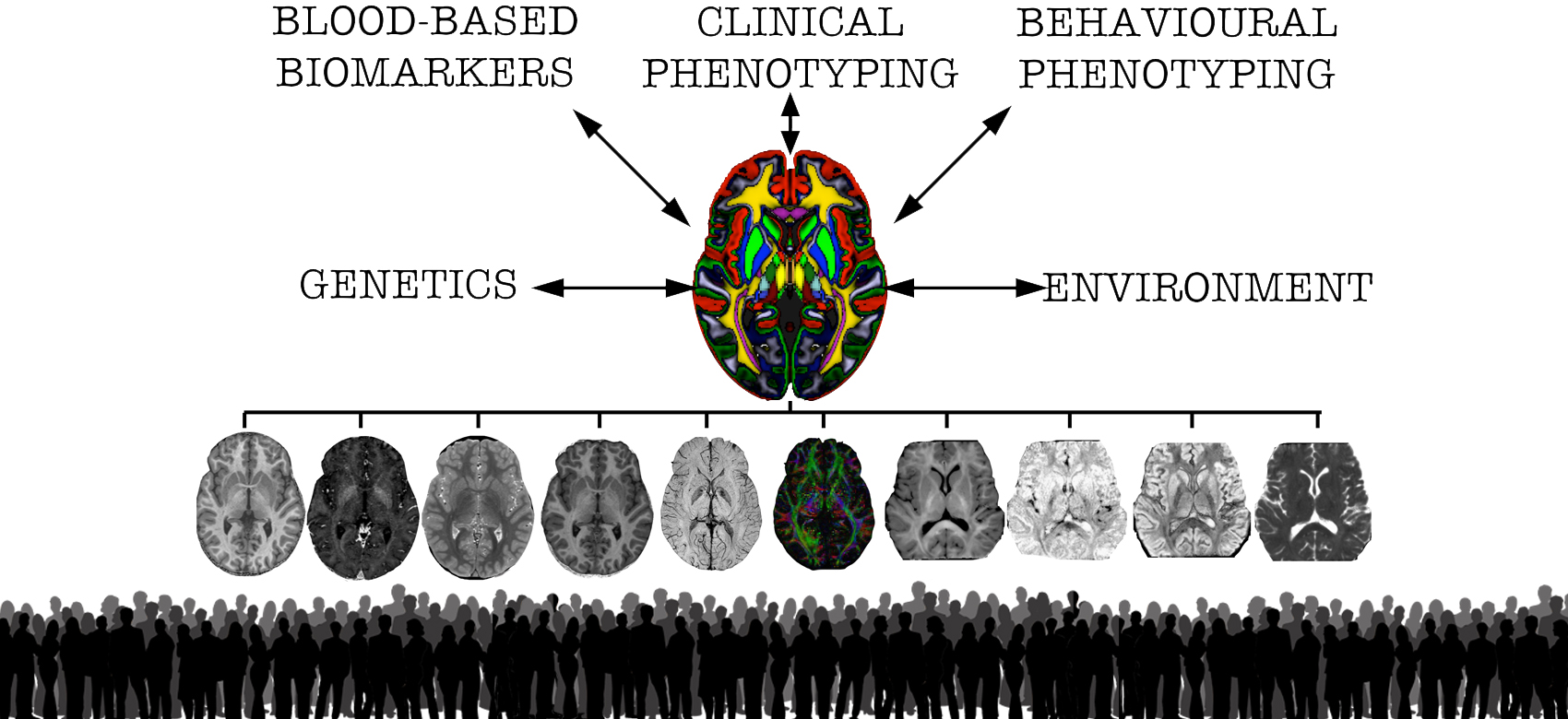
The causes of variability in brain function and disease between individuals are complex, with contributions from genetic, metabolic and environmental factors. The Anatomical Phenomics lab, lead by Dr Christian Lambert at the Functional Imaging Laboratory (FIL) in UCL, studies the link between differences in brain structure and inter-individual variability in health and disease. We develop and use advanced computational tools using quantitative MRI to enable in vivo MR-histology to study how differences in brain microstructure contributes to phenotypic variability. The over-arching aim is to better understand the mechanisms that dictate how a disease manifests and progresses at the level of the individual.
Improving our understanding of these relationships will allow:
- Earlier, more accurate diagnoses in conditions such as Parkinson’s disease (PD)
- Better predictions of future disease progression and prognosis
- More sensitive biomarkers, to track disease progression in the absence of clinical signs
- Insight into the biological foundations of structural-functional variation
Read about our current projects:
Quantitative MRI for Anatomical Phenotyping (qMAP): These are cohort studies that combine the advanced qMRI-based acquisitions and anatomical methods with detailed in-person assessments for deep phenotyping. The first, and largest, of these is focused on early stage Parkinson’s disease (PD) and related conditions (qMAP-PD). Initially funded by the UK Medical Research Council (MRC), and later extended via combined MRC and Parkinson’s UK funding, qMAP-PD currently consists of over 300 individuals, many with longitudinal follow-up extending up to seven years post-diagnosis. Other cohorts include Essential Tremor Syndrome (qMAP-ET, led by Dr Amit Batla), monogenic forms of Parkinson’s (mPD) and atypical Parkinson’s syndromes such as Multiple System Atrophy.
Multiscale Modelling in Parkinson’s disease: This is a “multiscale” extension to qMAP in collaboration with Professor Sonia Gandhi (Francis Crick Institute) and Professor Karl Friston (FIL, UCL) – In this, individual-specific induced Pluripotent Stem Cell (iPSC) models of cortical and midbrain neurons are being created for sPD and mPD to understand the cellular dynamics of disease and link these to changes in brain and phenotype seen in life, in the same individuals. This work embodies an emergent discipline that synergises molecular, cellular, clinical and computational neuroscience to better understand disease subtypes by their cause rather than their phenotype. Delivering tools to integrate data across scale in the same individuals, our vision is to provide an accessible framework, applicable to other disorders, to allow more rapid translation of targeted treatments that can be started before irreversible tissue loss, due to disease progression, has occurred